Introduction
The GlycoDomainViewer brings to the community an integrative tool for glycoproteomics that enables global analysis of the interplay between protein sequences, glycosites, types of glycosylation, and local protein fold / domain and other PTM context. The GlycoDomainViewer integrates poly-Omics data sets and enables exploration and visualisation of individual glycosites on proteins from many organisms. We provide a visual representation of protein context around glycosites, automatically managing the amount of information that is drawn so that data remains comprehensible. The tool aggregates and presents data produced experimentally as well as retrieved from other databases, with a view to presenting the maximum amount of relevant protein and post-translational modification (PTM) information available to explore the possible effects of glycosylation on a protein. In order to focus upon protein context, only initiating monosaccharides are shown, with limited glycan elaboration (i.e. T structures).
Usage
The following key can be used to interpret the symbols used in the GlycoDomain Viewer. Symbol notation follows the Symbol Nomenclature for Glycans scheme. Predictions from NetOGlyc4.0 have been pre-calculated for all Human proteins, and are shown whenever data is available.
Key
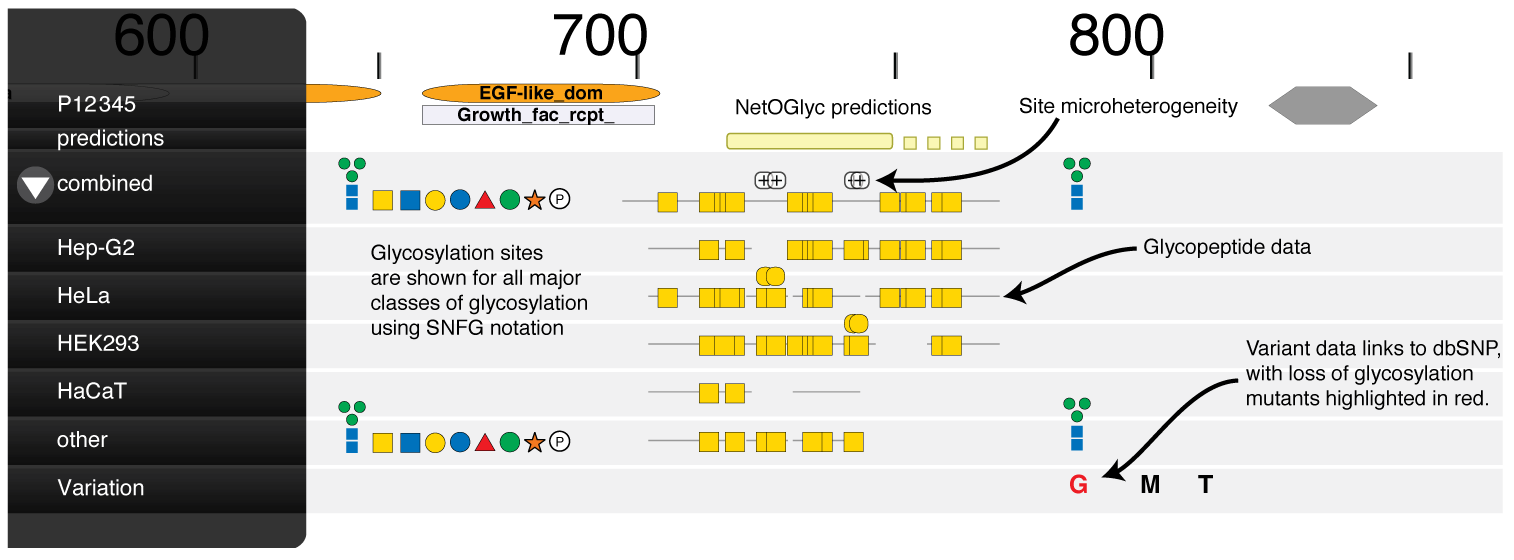
SNFG symbols
Citation
Steentoft, C. et al., 2013. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. The EMBO Journal, 32(10), pp.1478–1488
Included data
Data has been included from a wide variety of sources. Listed below are the papers that are currently being included in the GlycoDomainViewer.
Source Code
As usual, all source code for this site is available, with the main repository running at GitHub. If you're interested in using this software for your own purposes, get in contact with Hiren Joshi (hirenj) at GitHub.
